Digital skills and scholarship for researchers 5 – getting funded
Almost a year had past since that first conversation between Kaitlin Thaney, Nick Jones, Cameron McLean and myself where we asked:
‘what would Software Carpentry look like if it was delivered as a university course?’
A number of conversations and workshops were had that kept indicating that the thirst and need for this was there, that there wasn’t a clear solution in place, and that the solution was not going to be easy to produce. We knew what we wanted the house to look like, but we needed to find an architect. And of course, money to pay them.
Enter Nat Torkington
Nat organises an unconference called KiwiFoo. He invites a bunch of people to a retreat north of Auckland and lets the awesome happen. In 2015 I was invited, and, by pure luck Kaitlin Thaney was invited too as she was around that time in Australia for a software carpentry instructor training around ResBaz Melbourne. Also invited were Nick Jones, director of NeSi which had recently become the New Zealand institutional partner of Software Carpentry, and John Hosking, Dean of the Faculty of Science, University of Auckland.
The words that Kaitlin Thaney said at one of our meetings came back as if from a loudspeaker: You need to engage with the University Leadership. You need to think strategically.
And KiwiFoo gave us that opportunity.
Kaitlin, Nick and I brought John Hosking into the conversation, and his response was positive. We tried to exploit the convergence as much as we could over that weekend – there are not that many chances to get to sit with this group of people in a relaxed environment and without interruptions or the need to run to another meeting. We had each other’s full attention. And exploit we did.
Back in Auckland, Nick suggested that I talk about the project to the Centre of eResearch Advisory Board. The Centre of eResearch at the University of Auckland is helping researchers with exactly these kinds of issues. Next thing I know, Cameron McLean and I are trying to get everything we learned through the workshops into something more concrete. I talked to those details, and when the Board asked: ‘how can we help you’ I did not know what to say.
Dang.
Luckily, Nick Jones, as usual came to the rescue. We had a chat, and decided to work with me on higher level thinking. I was still missing the big picture that we could offer the leadership. Watching Nick’s thinking process was a humbling joy. I think I learned more from that session than what I did in all the Leadership programmes I was part of. I realised also how far I was from getting to where we needed to get. What is the long term vision? What are the gaps? Why do we need to fill them? How are you going to manage change?
At this meeting we saw we needed to engage with CLeaR, the organisation that provides Professional Development for staff and the group has a lot to offer in instructional design. We had already agreed that this training project should not be focused solely on students, but, rather, should have a broader scope. We produced an initial outline of what we were proposing, and invited Adam Blake from CLeaR to join the conversation and contribute to this document.
I was invited again to the eResearch Advisory Board, and this time I was better prepared. The timing was also perfect. The application window for the Vice Chancellor’s Strategic Development fund was open and I now knew what I needed: support to put an application through. We built a team of key project advisors, each who could contribute something quite specific: Adam Blake, to advise on course structure and to provide support to do the research on the course, Mark Gahegan, Director of the Centre for eResearch, Poul Nielsen, from the Auckland Bioengineering Institute, Nick Jones, from NeSI, and myself as the Project Lead, and the intention of hiring Cameron McLean as project manager. We worked on the application and backed, by the eResearch Advisory Board, it went in.
Our proposal was to develop a training suite, based on Software and Data Carpentry that could be used to be delivered to students and staff in different formats, to support a ResBaz in Auckland in February 2016, and to run a pilot course for students about to enter the research lab on second semester in 2016. We knew our bottleneck was time – people’s time to do the work. We asked for $150,000 in salaries.
In September we got the email: your application has been approved….
But…
The Vice Chancellor’s fund was giving us initially a limited amount of money with the rest of the money contingent on the approval of a needs analysis by the eResearch Advisory Board.
We accepted the offer and hired Cameron McLean as Project Manager (by now he was a trained Software Carpentry Instructor and had submitted his PhD thesis and was waiting for his viva). First order of business, a needs analysis.
Time to go to the library.
Digital skills and scholarship for researchers 4: the 3×3 table explained
When Billy Meinke and I sat to work on planning our sprint session for MozFest, he suggested that the activities of science could be grouped into 3 objects (text, code, data) and 3 actions (create, share, reuse). I was skeptical – surely science is way more complex than that. After running the session at MozFest and later in New Zealand, however, convinced me that Billy was right. We never encountered any object or action that we could not fit within that classification.
The actions:
The actions are self explanatory – create, share, reuse. You either are the author of a manuscript or you are not, you have contributed data or not, you have contributed to software or not. You create, share or reuse, or you don’t. However, what emerged at MozFest was how these 3 actions (which we seem to engage with apparently separately) are actually very dependent on each other – how we can share depends on how we created. Let’s look at an example:
I am capturing neural data using proprietary software that creates proprietary formats. That decision affects how those data can be reused (by a future self or others): only those with access to that software can open those files. Sharing and reuse, hence, becomes limited. If instead, I think of sharing and reuse from the onset, I may choose to use a piece of software (proprietary or not) that at least lets me export the raw data in an open format. Once I do this, then the opportunities to share for reuse come down to licencing. (Note: using a proprietary software may bring about other issues, but that is slightly a different discussion). So, during the act of creation we build constraints (or eliminate them) around sharing and reuse. So why not think about this upfront? Similarly, once we decide to share the licences we use will determine what kinds of reuse our work can have. These 3 actions are very interconnected – and it would be useful to think about how each affect each other from step 0 (planning). Those decisions will affect not just how we create, but also the infrastructure we choose to use so that the act of sharing and reuse is made as easy as possible when the time comes. Licences may state what we can do ‘legally’ but the infrastructure we use defines what we can do ‘easily’.
CC-BY Fabiana Kubke
The objects.
The objects became a lot more interesting as the MozFest and the New Zealand workshops progressed. At first, the idea of text, data and code seemed quite explanatory. Each of them we can identify with something we actually recognise, like a manuscript, the software we use for data acquisition and analysis, the data we measure or that is produced by some automated system. The fun part started when we tried to describe how these objects ‘behaved’, and, given those behaviours, how we were able to describe them (e.g., metadata).
Examples of text usually came across as manuscripts. When we think of manuscripts, we think of things that tell the story of data and code. They have a narrative that provides context to the work, there is usually a version of record which is difficult to modify, we usually publish it, it is peer reviewed, authors are well-defined, etc. The drafts prior to the version of record, the peer review, the corrections, etc., are usually not available (although that is changing in places, e.g., PeerJ, F1000, BioRxiv, to name a few). We usually interact with the versions of records; our ability to comment on those artefacts are limited , but are becoming available (e.g., PubMed) and mechanisms to suggest modifications (or to modify) these artefacts are almost non-existent. In other words – text (manuscripts) are stable. Another artefact that ‘behaves’ like text is equipment. Look at the following comparison:
| Manuscript | Equipment |
| Authors | Manufacturer |
| Article title | Model |
| DOI | Serial number |
| Volume, page numbers | Internal asset tag |
| Errata and corrections | Maintenance and repair records |
| etc | etc |
In other words, equipment seems to ‘behave’ like manuscripts. If you want to do (or say) something the equipment doesn’t do (or the paper doesn’t say) you need to buy (or write) a new one. So, when describing equipment you end up using similar descriptors to those for papers. In itself, a piece of equipment is a ‘stable’ object that can be described with ‘stable’ descriptors, not too dissimilar to a paper. Defective equipment breaks, defective papers get retracted. What this means is that the category ‘text’, when thought of as a set of behaviours and descriptors can help us build better descriptors for other artefacts with similar behaviours. This behaviour also determines how we create, share and reuse. How we publish a manuscript (behind paywalls, or with an Open Access licence) determines who can reuse and how. Using ‘open hardware’ equipment is similar to putting an open creative commons licence to a manuscript – using proprietary equipment is equivalent to publishing behind a paywall.
At the other end of the spectrum is code. Code likes to live in places like github. There is version control, the ability for multiple external and internal contributions, the list of contributors is agile and expands, and can stay alive and dynamic for long periods of time. There may be stable versions that are released at different time points, but in between, code changes. Code is at the core or reproducibility – it is the recipe that lists the ingredients and the sequence of what we did with and to the data. Not sharing the code is the equivalent of giving someone a cupcake and expecting them to be able to go and bake an identical version. So code is dynamic and its value is in the details. A lot of the value in code is that it is amenable to adaptations and modifications. One artefact of research that behaves like code is the experimental protocol, e.g., a protocol that describes a specific method for in situ hybridization, or how to make a buffer.
| Software | Protocol |
| Original author, plus future contributors | Original author plus future contributors |
| What a line of code does is described through annotation | What a line describing a step does ‘should’ be described by annotation |
| Bits and pieces can be copied to be part of another piece of software | Bits and pieces can be copied to be part of a different protocol |
| A single version for a single study | A single version for a single study |
| Otherwise constantly changing and being updated | Otherwise constantly changing and being updated |
| etc | etc |
So protocols seem to behave like code. Unfortunately, we tend to treat them as text (we share them in the materials and methods of our manuscripts). It would be much more useful to have protocols on places like github – allowing line testing and annotation, allowing ‘test driven development’ of protocols, allowing branching and merging, etc. If we were to think of protocols as ‘code’ we could then share them in a way that they could be more amenable for reuse. And if we do so, then we might think that an appropriate way to licence a protocol for sharing and reuse would be to apply the licences that promote sharing and reuse for code, not licences for text.
Data sits a bit in the middle of the two. Like text, it has stable versions – e.g., the data that accompanies a specific manuscript. Once data is captured, it cannot be changed (except of course to correct an error, or to legitimately remove ‘bad’ data points or outliers). In essence data changes by growing or reorganising, subsetting, etc, not by changing specific pre-existing values. It has some dynamic behaviours of code, and stable behaviours of text. It has stable versions and dynamic versions. How data is created determines how it can be shared and reused: are the formats open or proprietary, is it licenced openly or not, etc., as the project progresses, as outliers are eliminated, as new data is added. But for the most part there is a point in time where data moves from behaving like ‘code’ to behaving like text. Good open formats and licences can bring data back to a dynamic state (something harder to do with text-objects). This behaviour is important when we write the descriptors of data. There are the authors, data is linked to protocols and code, and eventually text, it can be used for different analysis, etc. Chemicals, in a way, behave like data:
| Data | Chemical |
| Author(s)/Contributors | Manufacturer |
| File name | Catalogue number |
| Version | Lot #, shipping date, aliquots |
| Storage place | Storage place |
| Linked to code and text | Linked to protocols and manuscripts |
| etc | etc |
How we share and describe data and chemicals is again similar. Is the chemical/data available to other researchers so they can repeat my experiments? Or is it something I produced in my lab and only share with a limited number of people? Again, how you ‘licence’ data and chemicals determines the extent to which these artefacts can be shared and reused. And, again, thinking about this intention at the planning stage makes a difference.
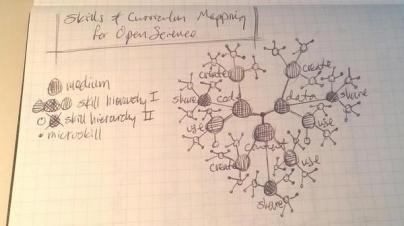
CC-BY Billy Meinke
All three objects can be published and cited, and data and code and slowly claiming the hierarchical position they deserve in the research cycle. The need for unique identifiers for resources is also recognised here and here, for example.
During the workshops it was fun to get people to ‘classify’ their research artefacts based on these behaviours. At MozFest, for example, Daniel Mietchen suggested his manuscripts behave more like code. I would argue that they should then be licenced (and described) like code.
What I learned from these workshops (and Billy’s 3×3 table) is that if we can classify all of our artefacts within these categories, then the process of describing our research artefacts and building them with the intention of openly sharing for reuse becomes much easier. And teaching the skills to understand how your choices constraint downstream effects becomes more achievable.
As long as, of course, you think about this from the beginning.
Footnote: This is my interpretation of Billy Meinke’s thinking model – he may loudly laugh about my interpretation. He may even roll his eyes, hit his head against the wall – I don’t know. But the clarity he brought to my approach to the problem is something I am extremely grateful for. Hat tip.
Open Access Week 2014
What do brain machine interfaces and Open Science have in common?
They are two examples of concepts that I never thought I would get to see materialised in my lifetime. I was wrong.

Kiwi Open Access Logo by the University of Auckland, Libraries and Learning Services is licensed under a Creative Commons Attribution 3.0 Unported License.
I had heard of the idea of Open Access as Public Library of Science was about to launch (or was in its early infancy) . It was about that time that I moved to New Zealand and was not able to go to conferences as frequently as I did in the USA, and couldn’t afford having an internet connection at home. Email communication (especially when limited to work hours) does not promote the same kind of chitter-chatter you might have as you wait in cue for your coffee – and so my work moved along, somewhat oblivious to what was going to become a big focus for me later on: Open Science.
About 6 years ofter moving to New Zealand things changed. Over a coffee with Nat Torkington, I became aware of some examples of people working in science embracing a more open attitude. This conversation had a big impact on me. Someone whom I never met before described me a whole different way of doing science. This resonated (strongly) because what he described were the ideals I had at the start of my journey; ideals that were slowly eroded by the demands of the system around me. By 2009 I had found a strong group of people internationally that were working to make this happen, and who inspired me to try to do something locally. And the rest is history.
What resonated with me about “Open Science” is the notion that knowledge is not ours to keep – that it belongs in the public domain where it can be a driver for change. I went to a free of fees University and we fought hard to keep it that way. Knowledge was a right and sharing knowledge was our duty. I moved along my career in parallel with shrinking funding pots and a trend towards academic commodification. The publish or perish mentality, the fears of being back-stabbed if one shares to early or too often, the idea of the research article placed in the “well-branded” journal, and the “paper” as a measure of one’s worth as a scientist all conspire to detract us from exploring open collaborative spaces. The world I walked into around 2009 was seeking to do away with all this nonsense. I have tried to listen and learn as much as I can, sometimes I even dared to put in my 2 cents or ask questions.
How to make it happen?
The biggest hurdle I have found is that I don’t do my work in isolation. As much as I might want to embrace Open Science, when the work is collaborative I am not the one that makes the final call. In a country as small as New Zealand it is difficult to find the critical mass at the intersection of my research interests (and knowledge) and the desire to do work in the open space. If you want to collaborate with the best, you may not be able to be picky on the shared ethos. This is particularly true for those struggling with building a career and getting a permanent position, the advice of those at the hiring table will always sound louder.
The reward system seems at times to be stuck in a place where incentives are (at all levels) stacked against Open Science; “rewards” are distributed at the “researcher” level. Open Research is about a solution to a problem, not to someone’s career advancement (although that should come as a side-effect). It is not surprising then how little value is placed in whether one’s science can be replicated or re-used. Once the paper is out and the bean drops in the jar, our work is done. I doubt that even staffing committees or those evaluating us will even care about pulling those research outputs and reading them to assess their value – if they did we would not need to have things like Impact Factors, h-index and the rest. And here is the irony – we struggle to brand our papers to satisfy a rewards system that will never look beyond its title. At the same time those who care about the content and want to reuse it are limited by whichever restrictions we chose to put at the time of publishing.
So what do we do?
I think we need to be sensitive to the struggle of those that might want to embrace open science, but are trying to negotiate the assessment requirements of their careers. Perhaps getting more people who embrace these principles at staffing and research University Committees might at least provide the opportunity to ask the right questions about “value” and at the right time. If we can get more open minded stances at the hiring level, this will go far in changing people’s attitudes at the bench.
I, for one, find myself in a relatively good position. My continuation was approved a few weeks ago, so I won’t need to face the staffing committee except for promotion. A change in title might be nice – but it is not a deal-breaker, like tenure. I have tried to open my workflow in the past, and learned enough from the experience, and will keep trying until I get it right. I am slowly seeing the shift in my colleagues’ attitudes – less rolling of eyes, a bit more curiosity. For now, let’s call that progress.
I came to meet in person many of those who inspired me through the online discussions since 2009, and they have always provided useful advice, but more importantly support. Turning my workflow to “Open” has been as hard as I anticipated. I have failed more than I have succeeded but always learned something from the experience. And one question that keeps me going is:
What did the public give you the money for?
ASAP Awards Finalists announced
(Cross-posted from Mind the Brain)
Earlier this year, nominations opened for the Accelerating Science Awards Program (ASAP). Backed by major sponsors like Google, PLOS and the Wellcome Trust, and a number of other organisations, this award seeks to “build awareness and encourage the use of scientific research — published through Open Access — in transformative ways.” From their website:
The Accelerating Science Award Program (ASAP) recognizes individuals who have applied scientific research – published through Open Access – to innovate in any field and benefit society.
The list of finalists is impressive, as is the work they have been doing taking advantage of Open Access research results. I am sure the judges did not have an easy job. How does one choose the winners?
In the end, this has been the promise of Open Access: that once the information is put out there it will be used beyond its original purpose, in innovative ways. From the use of cell phone apps to help diagnose HIV in low income communities, to using mobile phones as microscopes in education, to helping cure malaria, the finalists are a group of people that the Open Access movement should feel proud of. They represent everything we believed that could be achieved when the barriers to access to scientific information were lowered to just access to the internet.
The finalists have exploited Open Access in a variety of ways, and I was pleased to see a few familiar names in the finalists list. I spoke to three of the finalists, and you can read what Mat Todd, Daniel Mietchen and Mark Costello had to say elsewhere.
One of the finalist is Mat Todd from University of Sydney, whose work I have stalked for a while now. Mat has been working on an open source approach to drug discovery for malaria. His approach goes against everything we are always told: that unless one patents one’s discovery there are no chances that the findings will be commercialised to market a pharmaceutical product. For those naysayers out there, take a second look here.
A different approach to fighting disease was led by Nikita Pant Pai, Caroline Vadnais, Roni Deli-Houssein and Sushmita Shivkumar tackling HIV. They developed a smartphone app to help circumvent the need to go to a clinic to get an HIV test avoiding the possible discrimination that may come with it. But with the ability to test for HIV with home testing, then what was needed was a way to provide people with the information and support that would normally be provided face to face. Smartphones are increasingly becoming a tool that healthcare is exploring and exploiting. The hope is that HIV infection rates could be reduced by diminishing the number of infected people that are unaware of their condition.
What happens when different researchers from different parts of the world use different names for the same species? This is an issue that Mark Costello came across – and decided to do something about it. What he did was become part of the WoRMS project – a database that collects the knowledge of individual species. The site receives about 90,000 visitors per month. The data in the WoRMS database is curated and available under CC-BY. You can read more about Mark Costello here.
We’ve all heard about ecotourism. For it to work, it needs to go hand in hand with conservation. But how do you calculate the value (in terms of revenue) that you can put on a species based on ecotourism? This is what Ralf Buckley, Guy Castley, Clare Morrison, Alexa Mossaz, Fernanda de Vasconcellos Pegas, Clay Alan Simpkins and Rochelle Steven decided to calculate. Using data that was freely available they were able to calculate to what extent the populations of threatened species were dependent on money that came from ecotourism. This provides local organisations the information they need to meet their conservation targets within a viable revenue model.
Many research papers are rich in multimedia – but many times these multimedia files are published in the “supplementary” section of the article (yes – that part that we don’t tend to pay much attention to!). These multimedia files, when published under open access, offer the opportunity to exploit them in broader contexts, such as to illustrate Wikipedia pages. That is what Daniel Mietchen, Raphael Wimmer and Nils Dagsson Moskopp set out to do. They created a bot called Open Access Media Importer (OAMI) that harvests the multimedia files from articles in PubMed Central. The bot also uploaded these files to Wikimedia Commons, where they now illustrate more than 135 Wikipedia pages. You can read more about it here.
Saber Iftekhar Khan, Eva Schmid and Oliver Hoeller were nominated for developing a low weight microscope that uses the camera of a smartphone. The microscope is relatively small, and many of its parts are printed on a 3D printer. For teaching purposes it has two advantages. Firstly, it is mobile, which means that you can go hiking with your class and discover the world that lives beyond your eyesight. Secondly, because the image of the specimen is seen through the camera function on your phone or ipod, several students can look at an image at the same time, which, as anyone who teaches knows, is a major plus. To do this with standard microscopes would cost a lot of money in specialised cameras and monitors. Being able to do this at a relative low cost can provide students with a way of engaging with science that may be completely different from what they were offered before.
Three top awards will be announced at the beginning of Open Access Week on October 21st. Good luck to all!
Brain Hype
“Successful human-to-human brain interface” screamed the headlines – and so there I was clicking my way around the internet to read about it.
Those who know me also know that this is the kind of stuff what makes me tick, ever since learning about the pioneering work of Miguel Nicolelis. A bit over a decade ago I first heard of him, a Brazilian scientist working at Duke University in the Department where I spent a short tenure before moving to New Zealand. What I heard at the time was that he was attempting to extract signals from a brain and use them to control a robotic arm. I was quite puzzled by the proposition, I had been trained with the idea that each neuron in the brain is important and responsible of taking care of a specific bit of information. so thought I’d never get to see the idea succeed within my lifetime.
Nicolelis’ paradigm was relatively straightforward. He was to record the activity of a small area of the brain while the animal moved his arm, and identify what was going on in the brain during different arm movements. Activity combination A means arm up, combination B arm down, etc. He then would use this code to program a robotic arm so that the robotic it moved up when combination A was sent to it, down when combination B was sent, and so on. The third step was to connect the actual live brain to the robotic arm, and have the monkey learn that it had the power to move it himself.
What puzzled me at the time (and the reason that I thought his experiment couldn’t work) was that he was going to attempt to do this by recording the activity from what I could best describe as only a handful of neurons, and with rather limited control over the choice of those neurons. I figured this was not going to give him enough (or even the right) information to guide the movement of the robotic arm. But I was still really attracted to the idea. Not only did I love his deliberate imagination and how he was thinking outside the box,, but also, if he was successful, it would mean I’d have to start thinking about how the brain works in a completely different way.
It was not long before the word came out he had done it. He had managed to extract enough code from the brain activity that was going on during arm movements to program the robotic arm, and soon enough he had the monkey control the arm directly. And then something even much more interesting (at least to me) happened – the monkey learned that he could move the robotic arm without having to move his own arm. In other words, the monkey had ‘mapped’ the robotic arm into his brain as if it was his own. And that meant that it was time to revisit how I thought that brains worked.
I followed his work, and then in 2010 got a chance to have a chat with him at SciFoo. It was there that he told me how he was doing similar experiments but playing with avatars instead of real life robotic arms. how he saw this technology being used to build exoskeletons to provide mobility to paralyzed patients, and how he thought he was close to getting a brain to brain interface in rats.
A brain to brain interface?
Well, if the first set of experiments had challenged my thinking I was up for a new intellectual journey. Although by now I had learned my lesson.
I finally got to see the published results of these experiment earlier this year. Again, the proposition was straightforward. Have a rat learn a task in one room, collect the code and send that information to a second rat elsewhere and see if the second rat has been able to capture the learning. You can read more about this experiment from Mo Costandi here.
So when I heard the news about human to human brain interfaces, I inevitably got excited.
But then….
The paradigm of this preliminary study (which has not been published in a peer reviewed journal) is simple. One person is trying to play a video game imagining he pushes a firing button at the right time, and a second person elsewhere who actually needs to push the firing button for the game. The activity from the brain of the first person (this time recorded from the scalp surface) is transmitted to the brain of the second person through a magnetic coil (a device that is becoming commonly used to stimulate or inhibit specific parts of the brain.)
But is this really a bran to brain intterface?
Although the brain code of the first subject ‘imagining’ moving the finger was extracted (much like the Nicolelis group did back a decade ago), there is nothing about that code that is ‘decoded’ by the subject pressing the button. That magnetic coils can be used to elicit movement is not new. What part of the body moves depends on where on top of the head the coil is placed, and the type of zapping that is sent through the coil. So reading their description of the experiment, it seems that the signal that is being sent is a turn on/off to the coil, not a motor code in itself. The response from the second subject does not seem to need the decoding that signal – rather responding to a specific stimulation (not too unlike the kicking we do when someone tests our knee jerk reflex, or closing our eyelids when someone shines a bright light at our eyes).
I am also uncertain of how much the second subject knows about the experiment and I can’t help but wonder how much of the movement is self generated in response to the firing of the coil. Any awake person participating whose finger is put on top of a keyboard key and has a piece of metal on their head wouldn’t take too long to figure out how the experiment is meant to run.
There are a few comments (here and here for example) from readers identifying these weaknesses and even Nicolelis himself is quoted as saying it is too early to declare victory.
Which brings me back to the title of this post.
There is nothing wrong with sharing the group’s progress, In fact I think it is great, and I wish more of us were doing this. But I am less clear about what is so novel and what it contribute to our understanding of how the brain works to justify the hype.
This is a missed opportunity. There is value in their press release: here is a group that is sharing preliminary data in a very open way. This in itself is the news because this is good for science This should have been the hype.
Did you know?
- In 1978 a machine to brain interface (says Wikipedia) was successfully tested in a blind patient. Apparently progress was hindered by the patient needing to be connected to a large mainframe computer
- By 2006 a patient was able to operate a computer mouse and prosthetic hand using a brain machine interface that recorded brain activity using electrodes placed inside the brain. Watch the video.
- In 2009 using brain activity recorded from surface scalp electrodes to control a computer text editor, a scientist was able to send a tweet
References
- Carmena, J. M., Lebedev, M. A., Crist, R. E., O’Doherty, J. E., Santucci, D. M., Dimitrov, D. F., … Nicolelis, M. A. L. (2003). Learning to Control a Brain–Machine Interface for Reaching and Grasping by Primates. PLoS Biol, 1(2), e42. doi:10.1371/journal.pbio.0000042
- Pais-Vieira, M., Lebedev, M., Kunicki, C., Wang, J., & Nicolelis, M. A. L. (2013). A Brain-to-Brain Interface for Real-Time Sharing of Sensorimotor Information. Scientific Reports, 3. doi:10.1038/srep01319
- O’Doherty, J. E., Lebedev, M. A., Ifft, P. J., Zhuang, K. Z., Shokur, S., Bleuler, H., & Nicolelis, M. A. L. (2011). Active tactile exploration using a brain-machine-brain interface. Nature, 479(7372), 228–231. doi:10.1038/nature10489








leave a comment